
Oleg Mikhailovskii, Yi Xue, and Nikolai R. Skrynnikov.
https://doi.org/10.1107/S2052252521011891.
Read more »Tags: Mikhailovskii, Skrynnikov

Oleg Mikhailovskii, Yi Xue, and Nikolai R. Skrynnikov.
https://doi.org/10.1107/S2052252521011891.
Read more »Tags: Mikhailovskii, Skrynnikov

Dr. Boris B. Kharkov, Ivan S. Podkorytov, Dr. Stanislav A. Bondarev, Dr. Mikhail V. Belousov, Vladislav A. Salikov, Prof. Dr. Galina A. Zhouravleva, Prof. Dr. Nikolai R. Skrynnikov
https://doi.org/10.1002/anie.202102408
Read more »Tags: Kharkov, Podkorytov, Salikov, Skrynnikov
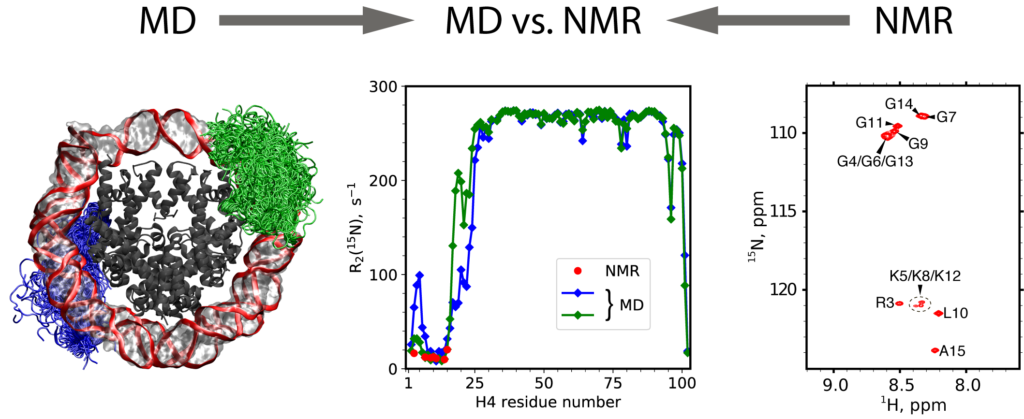
Sevastyan O. Rabdano, Matthew D. Shannon, Sergei A. Izmailov, Nicole Gonzalez Salguero, Mohamad Zandian, Rudra N. Purusottam, Michael G. Poirier, Nikolai R. Skrynnikov and Christopher P. Jaroniec
https://doi.org/10.1002/anie.202012046
Read more »Tags: Izmailov, Rabdano, Skrynnikov

Bolgov, A., Korban, S., Luzik, D., Zhemkov, V., Kim, M., Rogacheva, O., & Bezprozvanny, I. Crystal structure of the SH3 domain of growth factor receptor-bound protein 2. Acta Crystallographica Section F: Structural Biology Communications, 76(6), 263-270, 2020.
This study presents the crystal structure of the N-terminal SH3 (SH3N) domain of growth factor receptor-bound protein 2 (Grb2) at 2.5 Å resolution. Grb2 is a small (215-amino-acid) adaptor protein that is widely expressed and involved in signal transduction/cell communication. The crystal structure of full-length Grb2 has previously been reported (PDB entry 1gri). The structure of the isolated SH3N domain is consistent with the full-length structure. The structure of the isolated SH3N domain was solved at a higher resolution (2.5 Å compared with 3.1 Å for the previously deposited structure) and made it possible to resolve some of the loops that were missing in the full-length structure. In addition, interactions between the carboxy-terminal region of the SH3N domain and the Sos1-binding sites were observed in the structure of the isolated domain. Analysis of these interactions provided new information about the ligand-binding properties of the SH3N domain of Grb2.
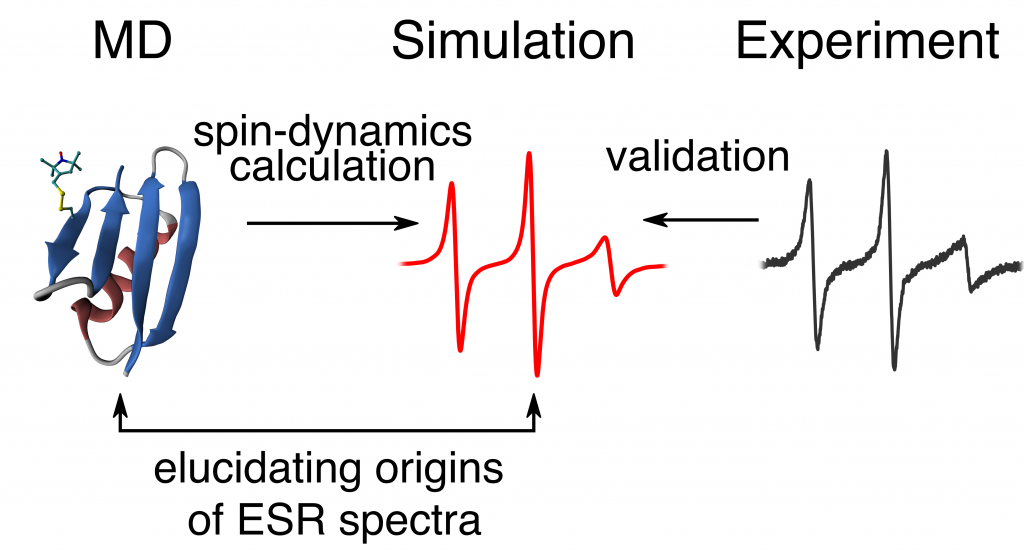
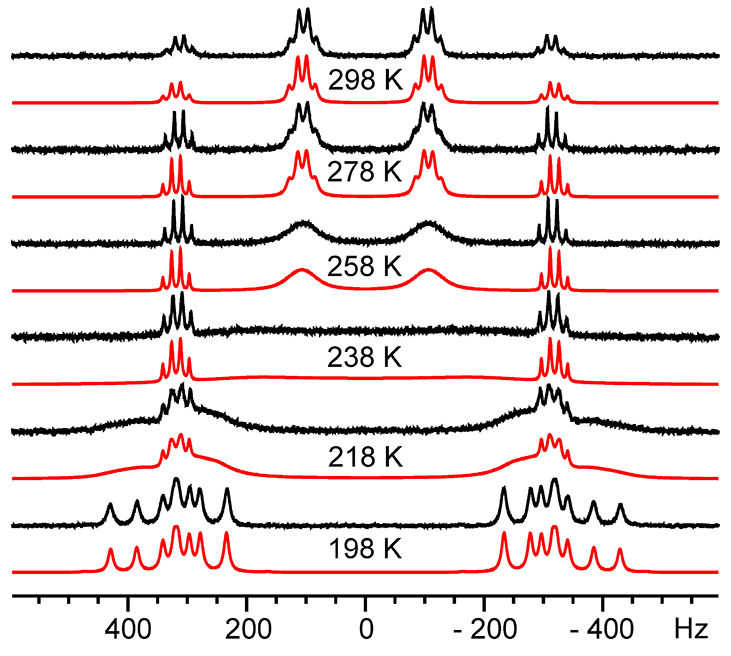
Izmailov, S.A, Rabdano, S.O., Hasanbasri, Z., Podkorytov, I.S., Saxena, S., Skrynnikov, N.R. Structural and dynamic origins of ESR lineshapes in spin-labeled GB1 domain: the insights from spin dynamics simulations based on long MD trajectories. Scientific Reports 2020.
DOI: 10.1038/s41598-019-56750-y
Tags: Izmailov, Podkorytov, Rabdano, Skrynnikov
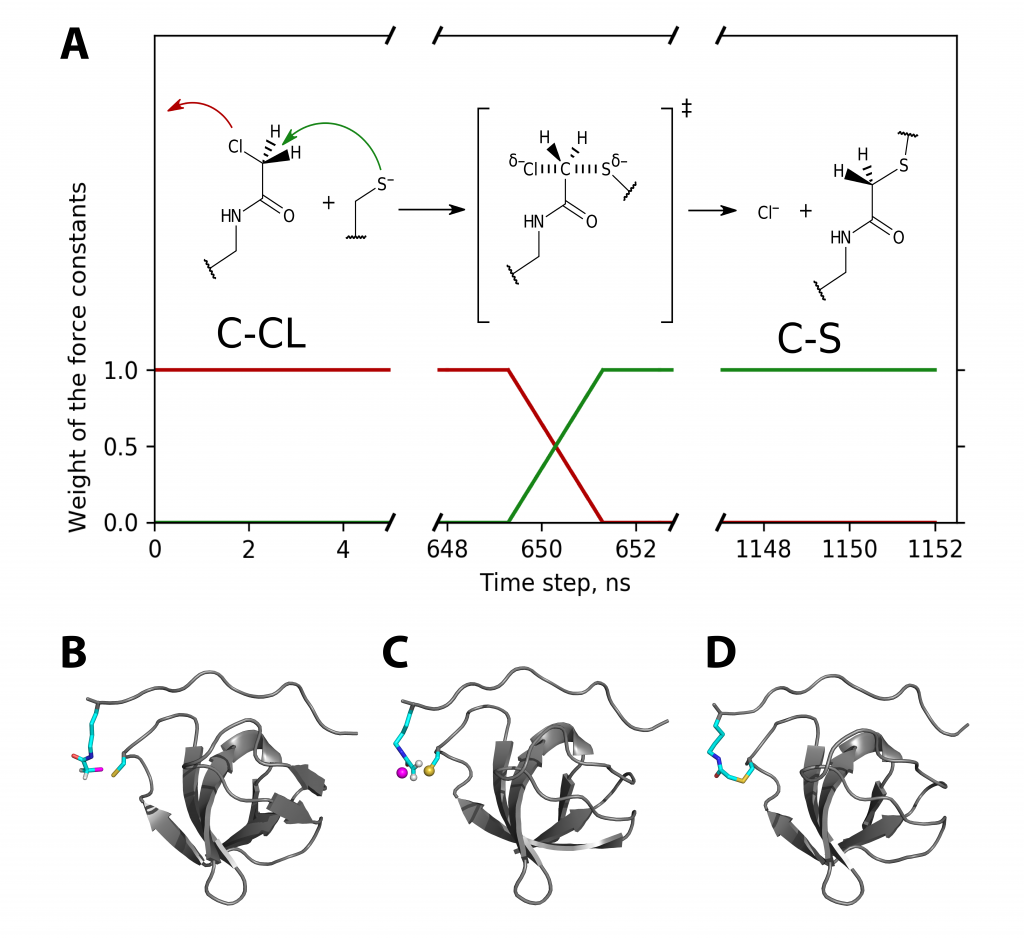
Luzik, D.A., Rogacheva, O.N., Izmailov, S.A., Indeykina, M.I., Kononikhin, A.S., Skrynnikov, N.R. Molecular Dynamics model of peptide-protein conjugation: case study of covalent complex between Sos1 peptide and N-terminal SH3 domain from Grb2. Scientific Reports 2019.
DOI:
Tags: Izmailov, Luzik, Rogacheva, Skrynnikov

Sevastyan Rabdano took part in the biophysical congress of European Biophysical Societies Association with the oral talk “Histones in nucleosome: fuzzy interaction with DNA”. The conference was held on July 20-24, 2019 in Madrid.
Tags: Rabdano
Titov, A. A., Filippov, O. A., Smol’yakov, A. F., Godovikov, I. A., Shakirova, J. R., Tunik, S. P., Podkorytov, I. S. & Shubina, E. S. Luminescent Complexes of the Trinuclear Silver(I) and Copper(I) Pyrazolates Supported with Bis(diphenylphosphino)methane. Inorg. Chem. 58, 8645–8656 (2019).
DOI: 10.1021/acs.inorgchem.9b00991
Tags: Podkorytov
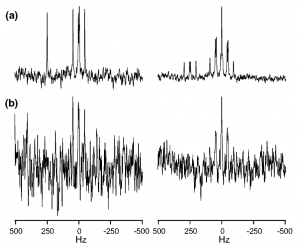
Podkorytov, I. S., Cherkasova, T. G., Kayfadzhyan, E. A. & Varshavsky, Yu. S. Multiplet-Matched Filtering of 103Rh Signal Using Information Contained in 31P Spectrum of AA′XX′ 31P–103Rh Spin System. Appl Magn Reson 50, 563–568 (2019).
DOI: 10.1007/s00723-018-1083-1
Tags: Podkorytov
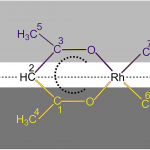
Varshavsky, Yu. S., Cherkasova, T. G., Galding, M. R., Korlyukov, A. A., Podkorytov, I. S., Gindin, V. A., Smirnov, S. N., Mazur, A. S. & Rubaylo, A. I. 13C NMR spectrum of crystalline [Rh(Acac) (CO)2]: A contribution to the discussion on [Rh(Acac) (CO)2] molecular structure in the solid state. Journal of Organometallic Chemistry 874, 70–73 (2018).
DOI: 10.1016/j.jorganchem.2018.08.009
Tags: Podkorytov