Nikolai Skrynnikov, Ivan Podkorytov, Sergei Izmailov, Olga Rogacheva, Kerstin Kaempf and Sevastyan Rabdano attended XXXVIII Finnish NMR Symposium, Jyvaskyla, Finland.
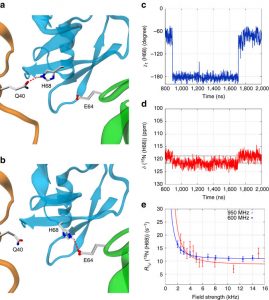
Nikolai Skrynnikov presented lecture “Dynamics in Protein Crystals: Insights From MD Simulations Complement New Solid-State NMR And X-Ray Data”.
Kerstin Kaempf presented oral talk “Local and global dynamics of intrinsically disordered proteins: a case study of H4 histone tail“.
Ivan Podkorytov presented poster “Diffusion-filtered experiment to detect flexible portion of protein chain in amyloid fibrils: application to Sup35NM“.
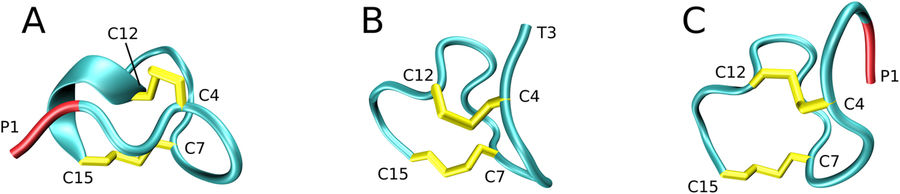
Sergei Izmailov presented poster “Simple MD-based model for oxidative folding of peptides and proteins”.
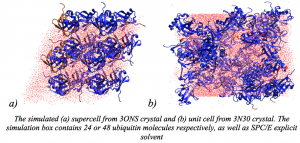
Olga Rogacheva presented poster “Using molecular dynamics simulation and chemical shift prediction to unravel dynamics in different crystal forms of ubiquitin”.
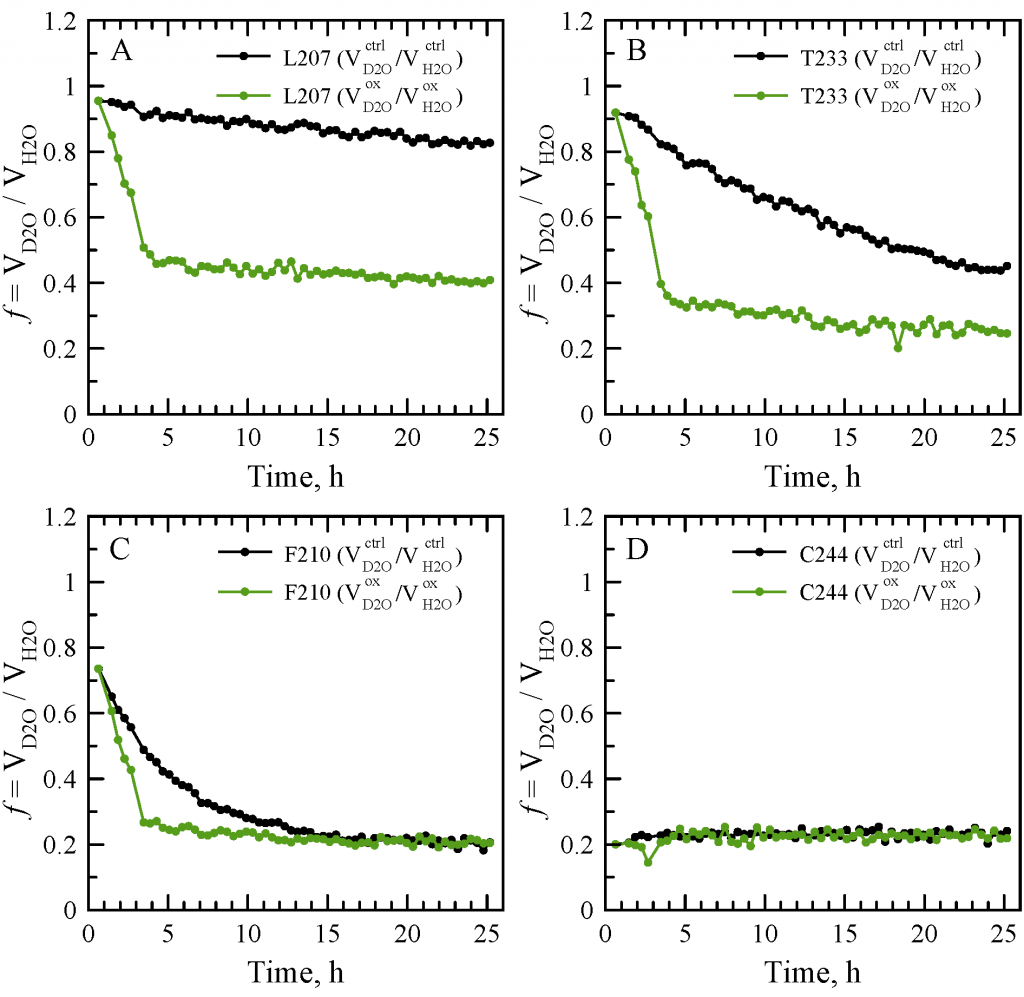
Sevastyan Rabdano presented poster “Loss of protein stability due to formation of intermolecular disulfide bonds under the effect of oxidative stress: case study of the RRM2 domain from neuropathological protein TDP-43”.
Tags: Finland, Izmailov, Jyvaskyla, Kaempf, Podkorytov, Rabdano, Rogacheva, Skrynnikov