Sevastyan O. Rabdano, Matthew D. Shannon, Sergei A. Izmailov, Nicole Gonzalez Salguero, Mohamad Zandian, Rudra N. Purusottam, Michael G. Poirier, Nikolai R. Skrynnikov and Christopher P. Jaroniec
https://doi.org/10.1002/anie.202012046
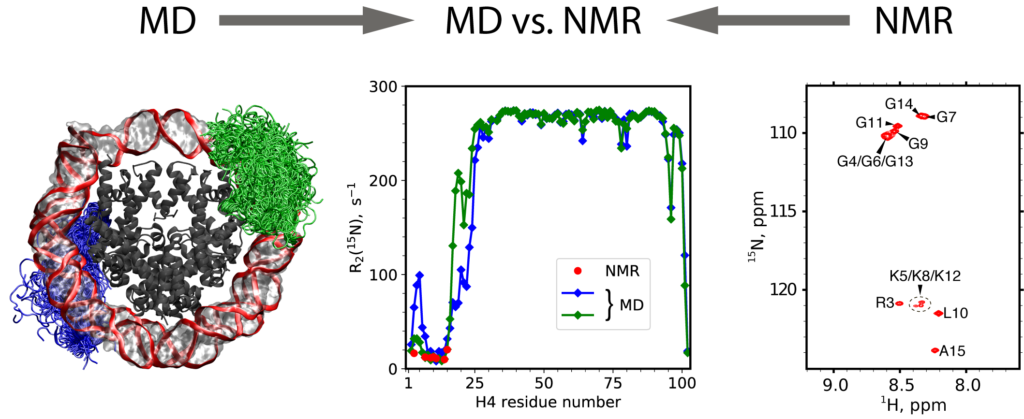
The interaction of positively charged N-terminal histone tails with nucleosomal DNA plays an important role in chromatin assembly and regulation, modulating their susceptibility to post-translational modifications and recognition by chromatin-binding proteins. Here, we report residue-specific 15N NMR relaxation rates for histone H4 tails in reconstituted nucleosomes. These data indicate that H4 tails are strongly dynamically disordered, albeit with reduced conformational flexibility compared to a free peptide with the same sequence. Remarkably, the NMR observables were successfully repro-duced in a 2-ms MD trajectory of the nucleosome. This is animportant step toward resolving an apparent in consistencywhere prior simulations were generally at odds with experimental evidence on conformational dynamics of histone tails. Our findings indicate that histone H4 tails engage in a fuzzy interaction with nucleosomal DNA, underpinned by a variable pattern of short-lived salt bridges and hydrogen bonds, which persists at low ionic strength (0–100 mM NaCl)
Tags: Izmailov, Rabdano, Skrynnikov
