
Tags: Skrynnikov, Vologda

Here, we present the new crystallography module xray, released as a part of the Amber 2023 package.
Oleg Mikhailovskii, Sergei A. Izmailov, Yi Xue, David A. Case, and Nikolai R. Skrynnikov
https://pubs.acs.org/doi/10.1021/acs.jcim.3c01531
Read more »Tags: Izmailov, Mikhailovskii, Skrynnikov
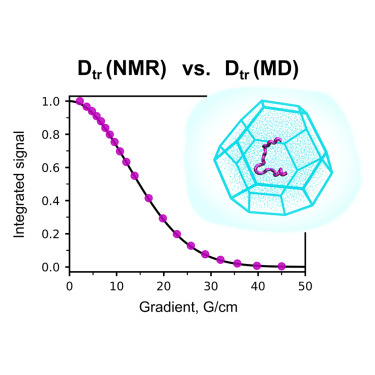
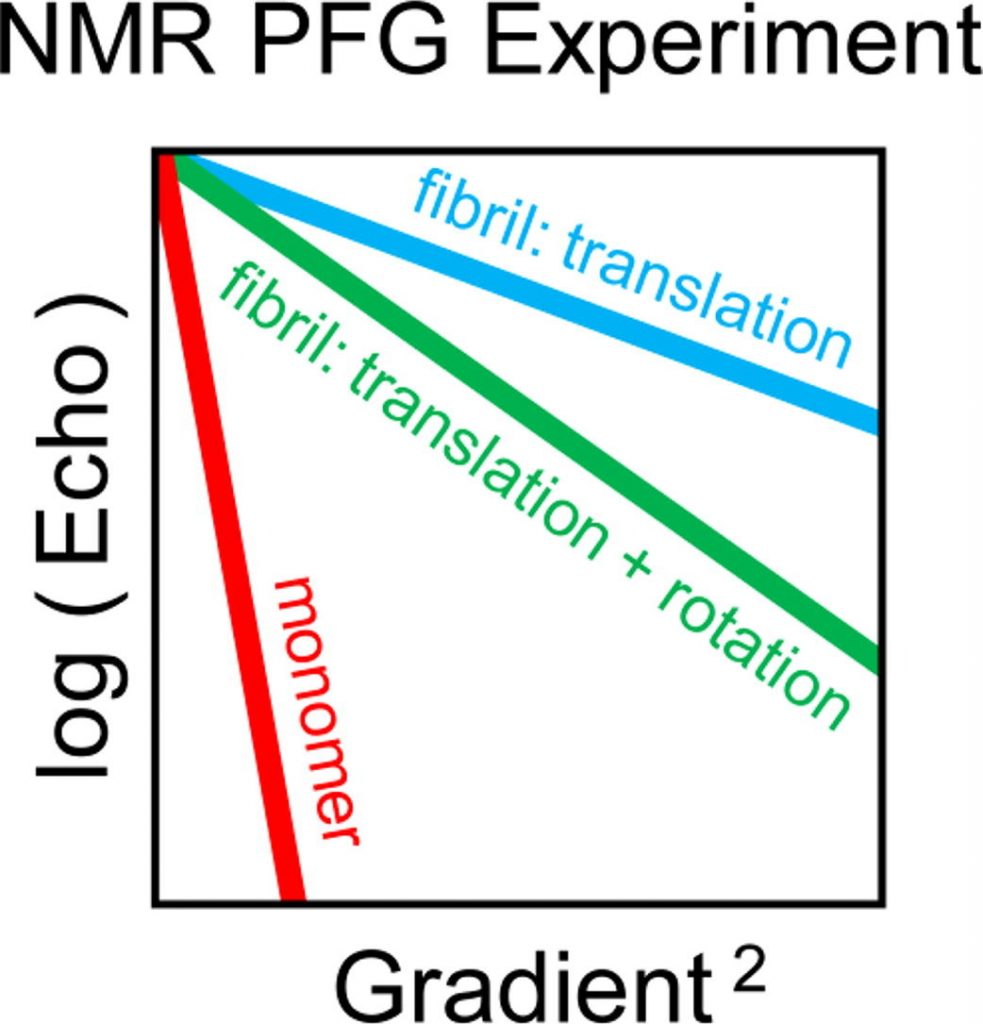
Olga O Lebedenko, Vladislav A Salikov, Sergei A Izmailov, Ivan S Podkorytov, Nikolai R Skrynnikov
https://www.sciencedirect.com/science/article/abs/pii/S0006349523007208
Read more »Tags: Izmailov, Podkorytov, Salikov, Skrynnikov

Wenjun Sun, Olga O. Lebedenko, Nicole Gonzalez Salguero, Matthew D. Shannon, Mohamad Zandian, Michael G. Poirier, Nikolai R. Skrynnikov, and Christopher P. Jaroniec
https://pubs.acs.org/doi/full/10.1021/jacs.3c10340
Read more »Tags: Lebedenko, Skrynnikov
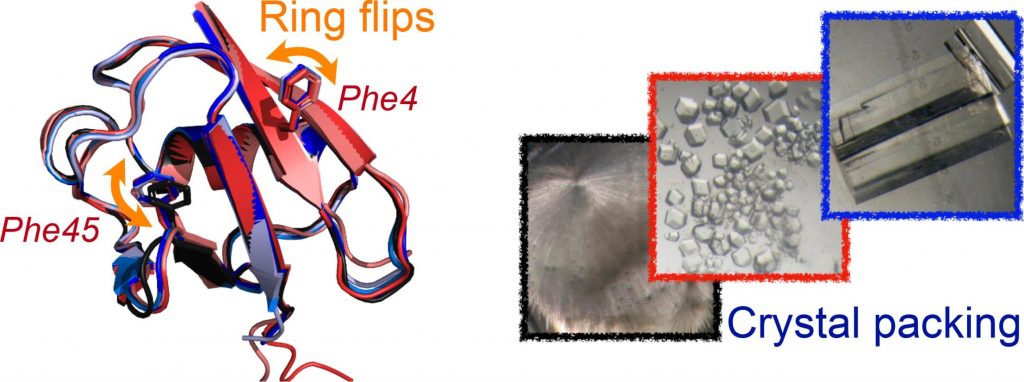
Diego F. Gauto, Olga O. Lebedenko, Lea Marie Becker, Isabel Ayala, Roman Lichtenecker, Nikolai R. Skrynnikov, Paul Schanda
https://www.sciencedirect.com/science/article/pii/S2590152422000204?via%3Dihub
Read more »Tags: Lebedenko, Skrynnikov
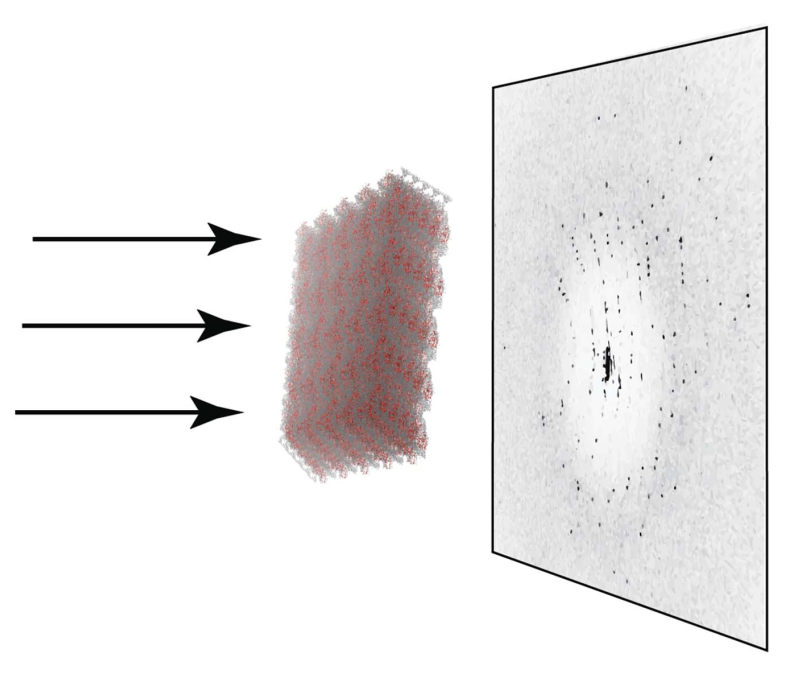
Ning Liu, Oleg Mikhailovskii, Nikolai R. Skrynnikovb, and Yi Xue
https://journals.iucr.org/m/issues/2023/01/00/lz5062/index.html
Read more »Tags: Mikhailovskii, Ning Liu, Skrynnikov, Yi Xue
Ivan S.Podkorytov, Nikolai R.Skrynnikov
https://doi.org/10.1016/j.jmr.2022.107303
Read more »
Tags: Podkorytov, Skrynnikov

Boris B. Kharkov
https://events.spbu.ru/events/translational-biomedicine-2022
Read more »Tags: Kharkov

D.A. Case, H.M. Aktulga, K. Belfon, I.Y. Ben-Shalom, J.T. Berryman, S.R. Brozell, D.S. Cerutti, T.E. Cheatham, III, G.A. Cisneros, V.W.D. Cruzeiro, T.A. Darden, R.E. Duke, G. Giambasu, M.K. Gilson, H. Gohlke, A.W. Goetz, R. Harris, S. Izadi, S.A. Izmailov, K. Kasavajhala, M.C. Kaymak, E. King, A. Kovalenko, T. Kurtzman, T.S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, M. Machado, V. Man, M. Manathunga, K.M. Merz, Y. Miao, O. Mikhailovskii, G. Monard, H. Nguyen, K.A. O’Hearn, A. Onufriev, F. Pan, S. Pantano, R. Qi, A. Rahnamoun, D.R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, A. Shajan, J. Shen, C.L. Simmerling, N.R. Skrynnikov, J. Smith, J. Swails, R.C. Walker, J. Wang, J. Wang, H. Wei, R.M. Wolf, X. Wu, Y. Xiong, Y. Xue, D.M. York, S. Zhao, and P.A. Kollman
https://ambermd.org/doc12/Amber22.pdf
Read more »Tags: Izmailov, Mikhailovskii, Skrynnikov, Yi Xue
B. Kharkov, X. Duan, J. Rantaharju, M. Sabba, M.H. Levitt, J.W. Canary, A. Jerschow
https://pubs.rsc.org/en/content/articlelanding/2022/cp/d1cp05537b
Read more »Tags: Kharkov